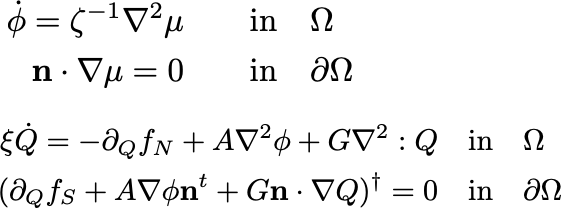RESEARCH
How do cells form embryos and tissues?
Embryos and tissues are complex systems composed of interacting, autonomous agents—the cells, that have evolved to assemble robustly into tissues and organs.
Despite major advances in molecular genetics and live imaging, we still understand little about how a single cell gives rise to a structured embryo or tissue. Our research explores fundamental questions such as:
“– How do cells control their shape, position, and fate?
– What information does a cell process to make decisions?
– How is morphogenetic information transmitted across the embryo?
– What principles ensure robustness in early development?”
To address these questions, we pursue a three-pronged strategy:
Integrating scales: We investigate how spatial and temporal scales interact during early embryogenesis and tissue morphogenesis. This multiscale approach is essential to connect predictive models with quantitative experimental data.
Reverse-engineering feedbacks: We develop new methods to infer minimal feedback loops between biochemical signaling and mechanics directly from data. These insights are then incorporated into realistic 3D simulations to capture multicellular self-organization.
Comparative embryology meets physics: We study a diverse range of embryos—from mammals to nematodes, ascidians, and spiralians—to uncover conserved and divergent principles. Through this comparative approach, we aim to help shape the emerging field of evolutionary biophysics.
1. Mechanobiology across scales
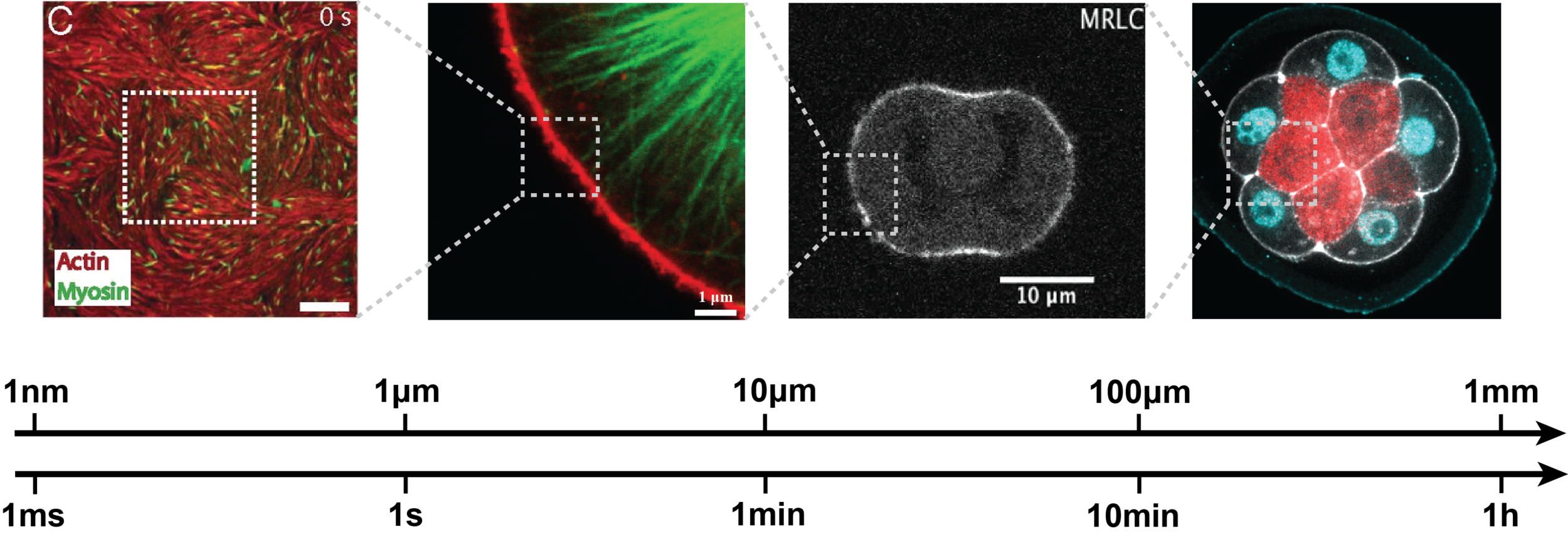
Morphogenesis spans multiple spatial and temporal scales, while most perturbation tools act at the molecular level.
Bridging this gap requires understanding how molecular players shape the physical properties of cells—a major experimental and theoretical challenge. Below are examples of our research addressing this question.
MEasuring
In close collaboration with biologists and experimental physicists we are developing or improving methods to probe cell and tissue mechanics, including AFM (Atomic Force Microscopy), tissue compression and mechanical inference from microscopy images.
Publications:
Ichbiah et al, Nature Methods 2023
Yamamoto et al, Developmental Cell 2025
Delbary et al., in preparation 2025.
Collaborations:
Guillaume Charras (University College London, UK)
Charles Baroud (Institut Pasteur, FR),
Myriam Reffay (Université Paris Cité, FR)
Modeling
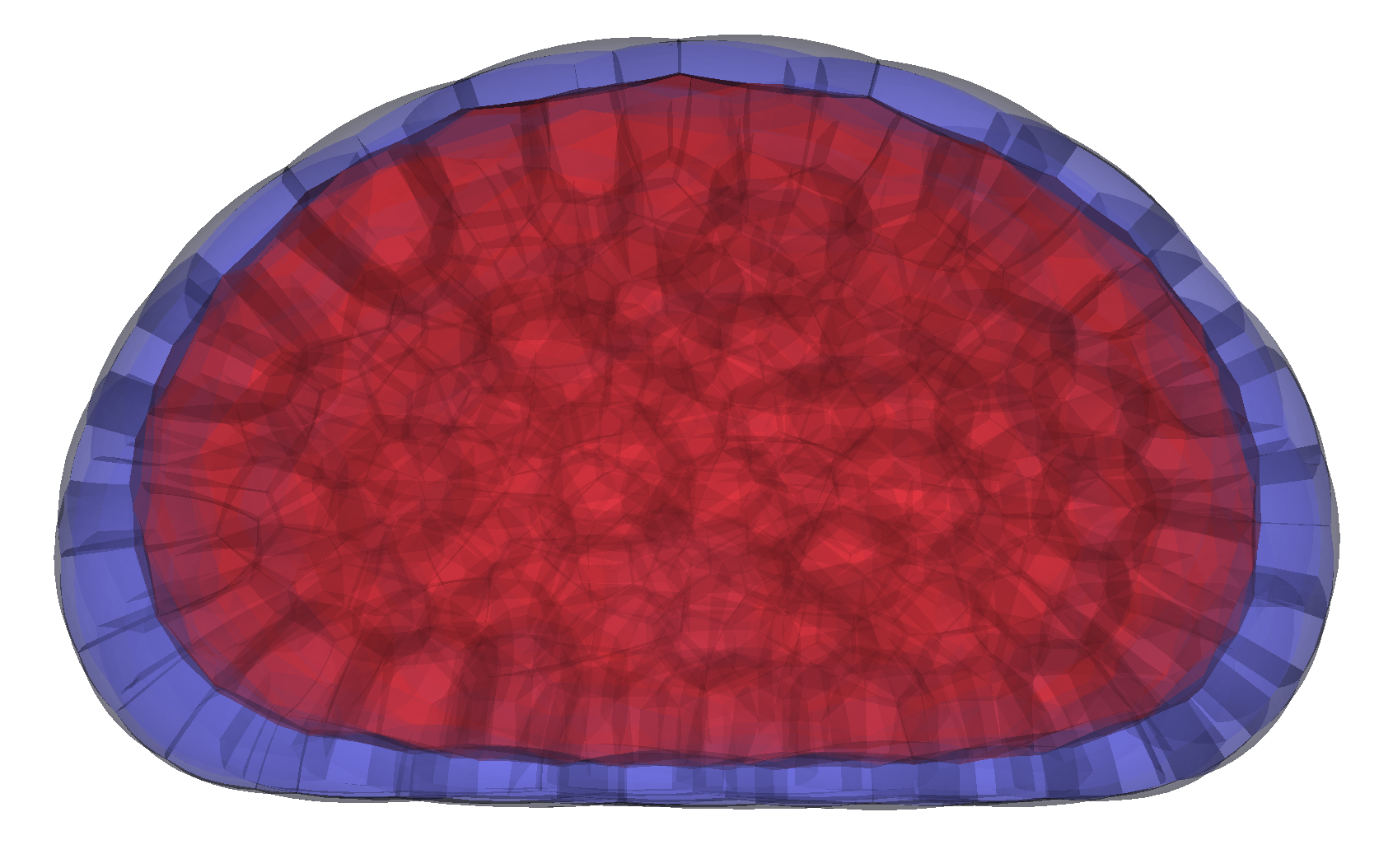
We develop theoretical models and computational tools to predict how active forces shape biological structures, from single cells to whole tissues. This includes modeling membrane–cortex interactions, osmo-hydraulic control of morphogenesis, the active nematic behavior of cell surfaces and 3D tissues, as well as discrete-geometric and novel finite-element methods for 3D simulations.
Publications:
Le Verge-Serandour & Turlier, PLoS Comp Biol 2021
Borja, Bleyer & Turlier J Mecha Phys Solids 2022
Neiva & Turlier, submitted to JCP 2025
Le Verge-Serandour & Turlier, in preparation
Predicting
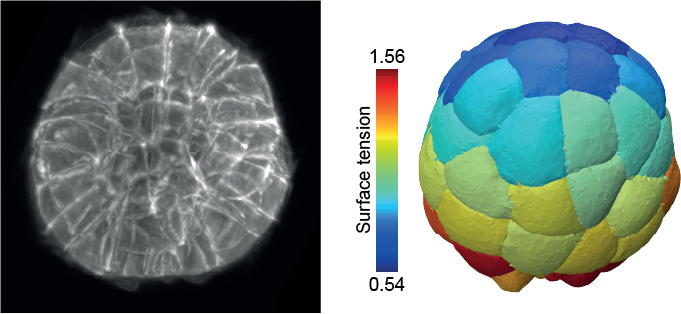
Our physical models enable us to predict how molecular players drive shape formation across scales—from subcellular structures to whole embryos and tissues—in close collaboration with biologists. These models have helped uncover key morphogenetic mechanisms, such as compaction and blastulation in the mammalian embryo, spindle (de)centering mechanisms, and the transmission of membrane tension across the cell.
Publications:
Dumortier et al., Science 2019
de Belly et al., Cell 2023
Rosfelter et al., J Cell Science 2024
Firmin et al., Nature 2024
Collaborations:
Alex McDougall & Rémi Dumollard (Villefranche-sur-mer, FR),
Ana Carvalho (i3S University of Porto, PT)
Orion Weiner (University of California San Francisco, US)
Nicolas Rivron (Institute of Molecular Biotechnology, AT)
2. Artificial Intelligence for morphogenesis
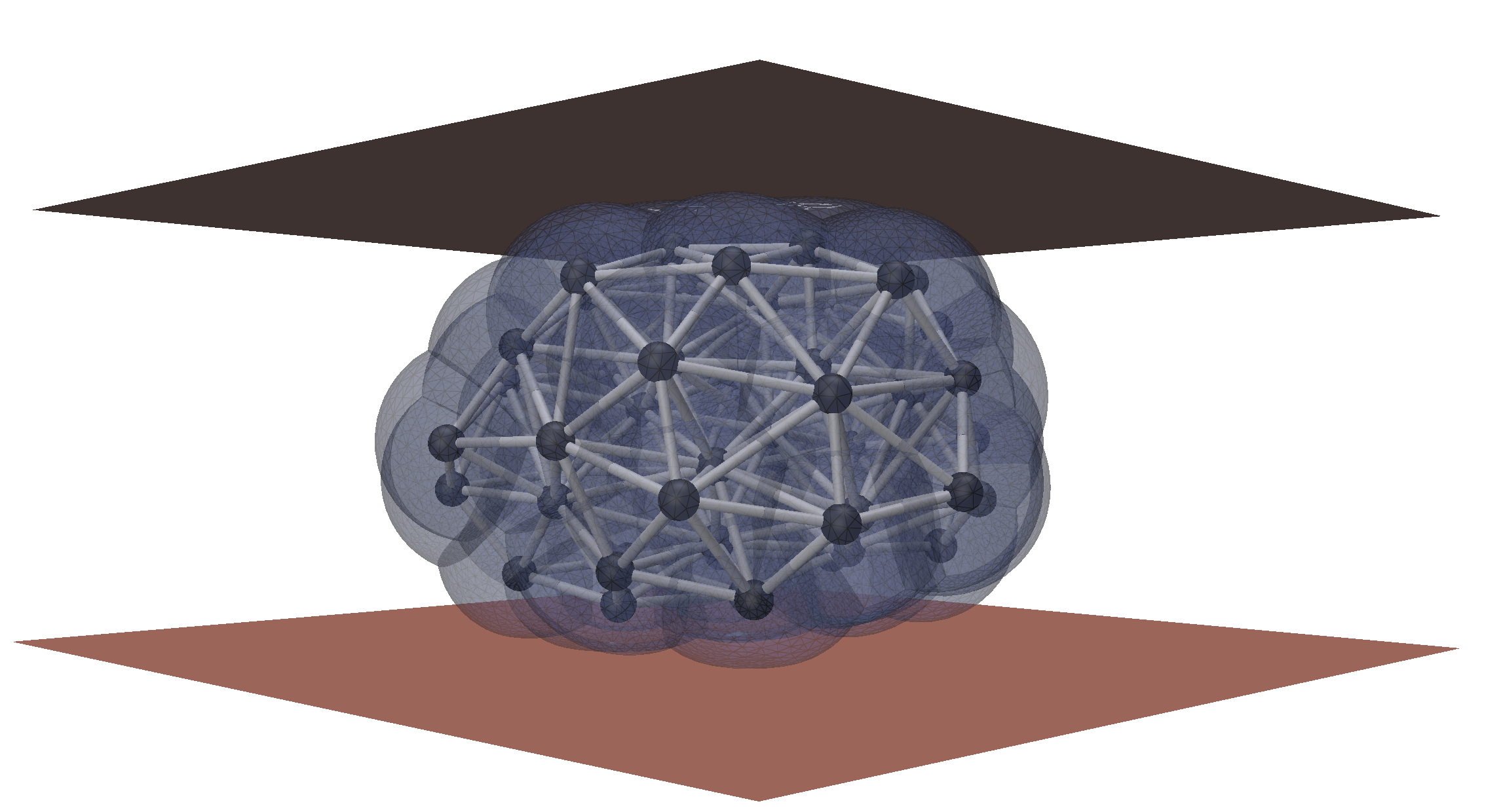
image Analysis and Inverse modeling
We develop automated tools for microscopy image analysis and the extraction of physical information. Our methods rely on differentiable modeling to directly link physical models with 3D images by solving inverse problems.
Publications:
Ichbiah et al., accepted to ICCV 2025
Pasqui et al., in preparation
Collaborations:
Maxence Ernoult (DeepMind, Paris FR)
Graph neural neworks for tissue modeling
We are developing a Graph Neural Network (GNN) surrogate model of tissue, enabling large-scale simulations of 3D foam-like tissue morphogenesis with integrated biochemical signaling.
Publications:
Goomanee & Turlier, in preparation
Reinforcement learning for morphogenesis
IN CONSTRUCTION
Publications: